Advanced Network Analysis
Community Detection
Olga Chyzh [www.olgachyzh.com]
Network Community Detection
Reading Materials
Barabasi, Albert-Laszlo. (2016). Network Science. Cambridge University Press. Chapter 9.
Zachary, Wayne W. (1977). An information flow model for conflict and fission in small groups. Journal of Anthropological Research 33(4) : 452-473.
Renshon, Jonathan. (2016). Status deficits and war. International Organization 70(3): 513-550.
Gould, Roger V. (1991). Multiple Networks and Mobilization in the Paris Commune, 1971. Americal Sociological Review 56(6): 716-729.
Cruz, Cesi, Julien Labonne, and Pablo Querubin. (2020). Social network structures and the politics of public goods provision: evidence from the Philippines. American Political Science Review 114(2): 486–501
Introduction: Belgium
Introduction: Belgium
Population: 11.5 mil
Bilingual: 59% Flemish (speak Dutch), 40% Waloons (French)
Is the society so densely knitted together that nobody notices who is of what ethnic group?
Or do the two groups minimize the interactions?
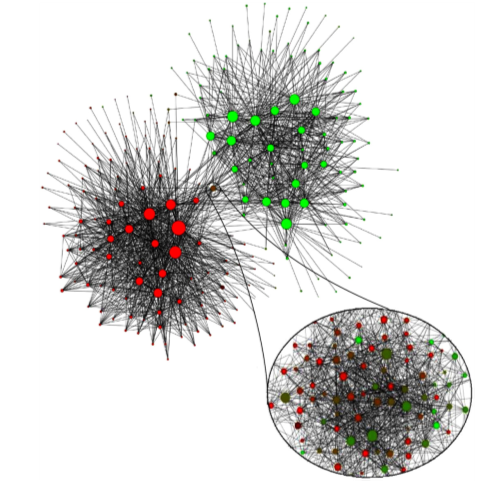
Blondel et al. (2008)
Applied a community finding algorithm to the call patterns of a big mobile phone operator.
Goal: identify individuals who regularly talk to one another.
Blondel et al. (2008)
Communities: the Basics
Network Levels of Analysis

What Is A Community?
A community is a locally dense connected subgraph. (Barabasi, 2016, 325)
All members of a community must be reached through other members of the same community (connectedness).
Nodes that belong to the same community have a higher probability of linking than nodes outside of the community (density).
Examples: communities among the karate club members, communities of international states, communities of legislators, neighborhood communities.
Note that these features do not uniquly define a community, just offer some guidelines.
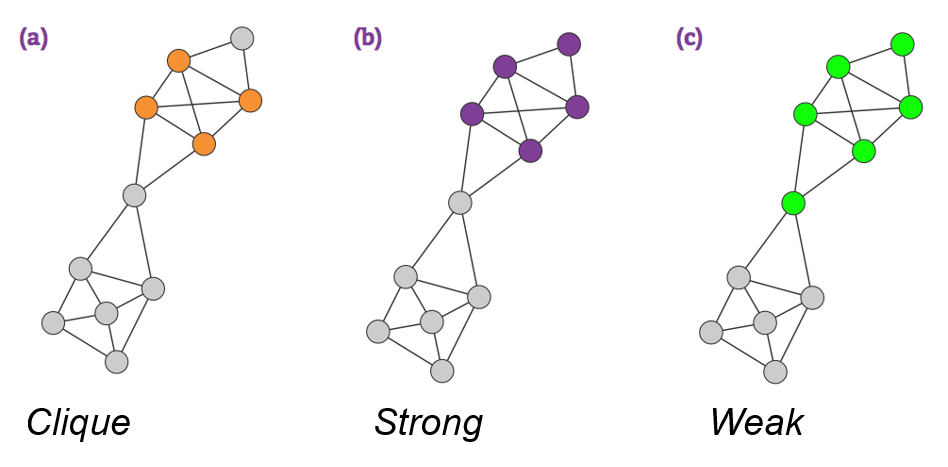
Cliques as Communities
A clique is a complete subgraph of k-nodes.
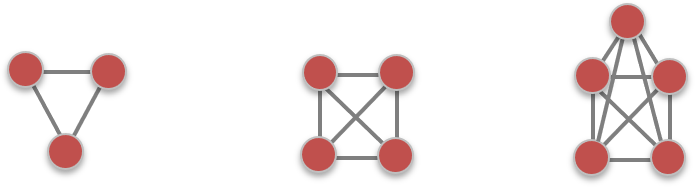
- May be too restrictive.
Strong and Weak Communities
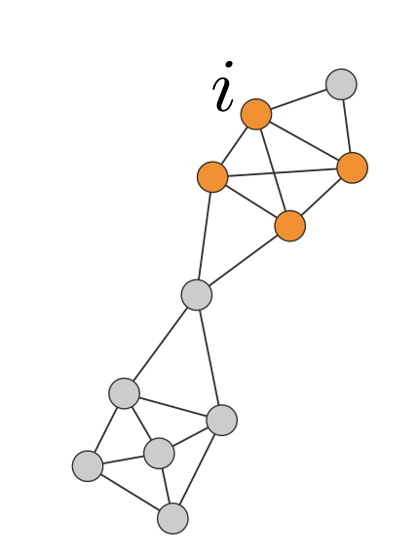
Consider a connected subgraph C of Nc nodes.
Internal degree, kinti is the set of links of node i that connectes to the other nodes in the same community.
External degree, kexti is the set of links of node i that connects to the rest of the network.
If kexti=0, all neighbors of i belong to C, and C is a good community for i.
If kinti=0, all neighbors of i belong to other communities, then i should be assigned to a different community.
Example
kexti=1, kinti=3
Strong and Weak Communities
In a strong community each node of C has more links within the community than with the rest of the graph, kinti(C)>kexti(C). In a weak community, the total internal degree of C exceeds its total external degree, kin(C)>kout(C).
Number of Partitions
How many ways can we partition a network into 2 communities?
Divide a network into two equal non-overlapping subgraphs, such that the number of links between the nodes in the two groups is minimized.
Two subgroups of size n1 and n2. Total number of combinations: N!n1!n2!
- N=10⟹256 partitions
- N=100⟹1026 partitions
If the number and size of the communities are unknown at the beginning, the number of possible partitions is a Bell Number.
Brute force approach is unfeasible, need an algorithm.
Zachary (1977)
Karate club of 34 members;
78 pairwise links between members who regularly interacted outside the club;
A conflict between the club’s president and the instructor split the club into two.
Today community finding algorithms are often tested based on their ability to infer these two communities from the structure of the network before the split.
Zachary (1977)
- Maximum Flow Minimum Cut Procedure: divides network nodes into two subsets, one containing the source and the other, the sink. The edges connecting the two subsets, called the cut. The cut with the minimum sum of the capacities of all of its edges represents a bottleneck in the network. (464)
Zachary (1977)
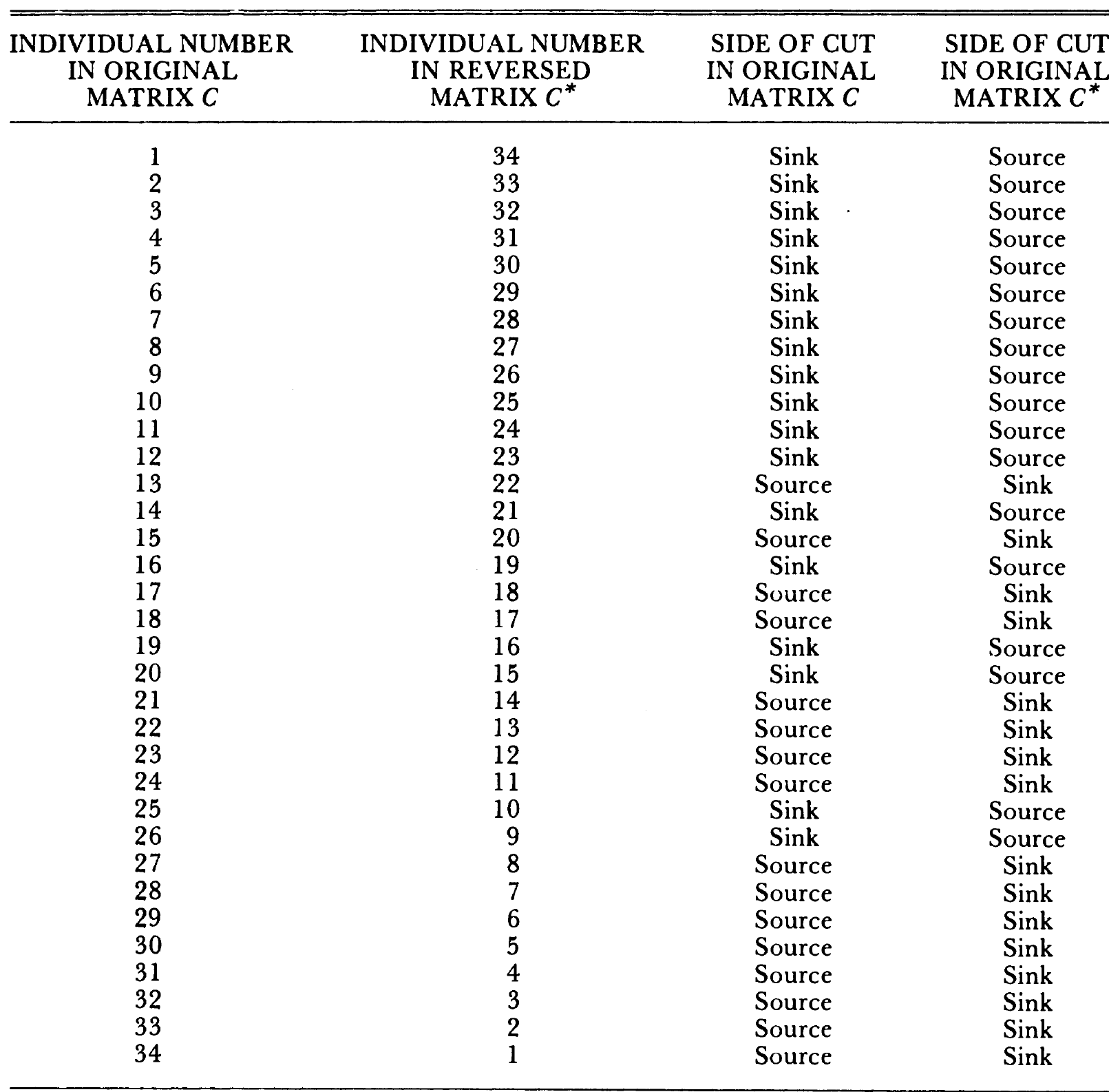
Zachary (1977)
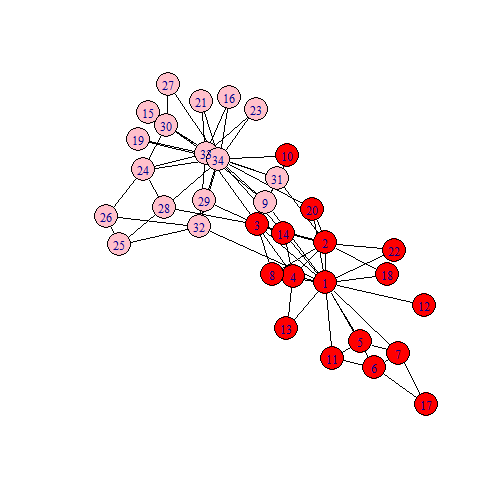
Zachary (1977)
mf$partition1## + 17/34 vertices, from 63d649f:## [1] 1 2 3 4 5 6 7 8 10 11 12 13 14 17 18 20 22mf$partition2## + 17/34 vertices, from 63d649f:## [1] 9 15 16 19 21 23 24 25 26 27 28 29 30 31 32 33 34Other Ways to Partition Networks
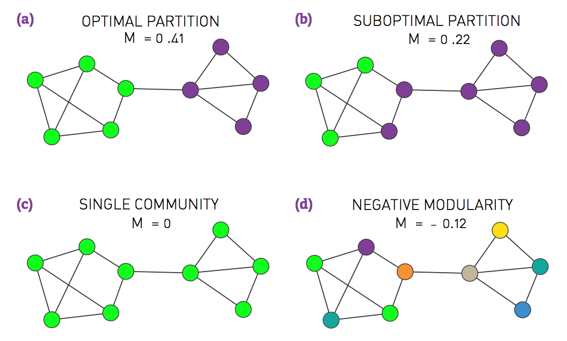
- Modularity: a measure of the extent to which like is connected to like in a network.
- A greedy algorithm iteratively joins nodes if the move increases the new partition's modularity.
The Girvan–Newman algorithm
Used in Cruz et al 2020
Calculate betweenness for all edges in the network
Remove the edge with the highest betweenness
Recalculate betweenness for all edges affected by the removal
Repeat from step 2 until no edges remain
From the resulting dendrogram (the hierarchical mapping produced by gradually removing these edges), select the partition that maximizes network modularity
Walktrap algorithm
Relies on the idea that random walks on a graph tend to get “trapped” into densely connected parts corresponding to communities.
The algorithm a large number of random walks and groups together nodes that are tied together through those walks
See Pons and Latapy (2005)
Comparing Clustering Algorithms
Lab
Zachary (1977)
library(igraph) #Zachary's karate club dataset is built into the igraph package.karate <- make_graph("Zachary") #loads the data from the igraph packagemf<-max_flow(karate, source=V(karate)[1], target=V(karate)[34])V(karate)$color<- ifelse(V(karate) %in% mf$partition1, "red","pink")plot(karate, edge.color="black", vertex.frame.color="black")mf$partition1mf$partition2library(sna)data(coleman)#make into an igraph objectfriends<-graph_from_adjacency_matrix(coleman[2,,], mode="undirected", diag=FALSE)friends <- igraph::delete.vertices(friends , which(igraph::degree(friends)==0))LO = layout_with_fr(friends) #Layoutcfg<-cluster_fast_greedy(friends)modularity(cfg)## [1] 0.5904201cfg$membership## [1] 5 5 1 1 5 5 5 5 5 1 5 5 5 5 1 5 5 5 5 5 5 3 5 1 4 5 1 4 4 1 1 1 4 1 4 5 1 4## [39] 1 1 1 2 2 2 2 2 2 2 2 2 3 2 2 3 2 3 2 3 3 3 3 3 3 3 3 3 3 3 3Communities in the Friendship Network
plot(cfg, friends, layout=LO, main="Greedy")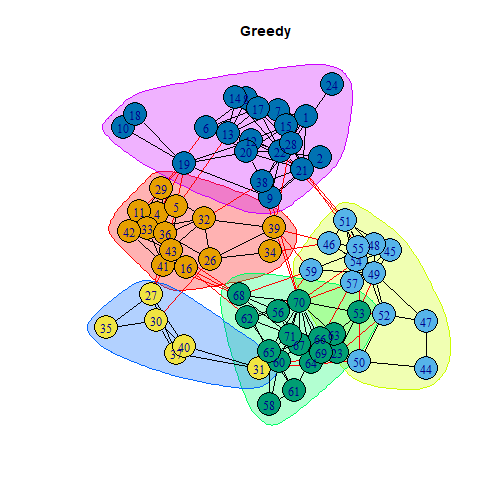
Other Examples:
wc <- cluster_walktrap(friends) #community structure via short random walksmodularity(wc)## [1] 0.5935815membership(wc)## 1 2 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 26 27 28 ## 2 2 1 1 2 2 2 2 1 1 2 2 2 2 1 2 1 1 2 2 2 3 2 2 5 2 ## 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 ## 1 5 5 1 1 2 5 1 5 2 2 5 1 1 1 3 3 3 3 3 3 3 3 3 3 3 ## 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 ## 3 4 3 4 3 4 4 4 4 4 4 4 4 4 4 4 4Fast Greedy Algorithm
fg <- cluster_fast_greedy(friends) #Used in Renshon (2016)modularity(fg)## [1] 0.5904201fg$membership## [1] 5 5 1 1 5 5 5 5 5 1 5 5 5 5 1 5 5 5 5 5 5 3 5 1 4 5 1 4 4 1 1 1 4 1 4 5 1 4## [39] 1 1 1 2 2 2 2 2 2 2 2 2 3 2 2 3 2 3 2 3 3 3 3 3 3 3 3 3 3 3 3Egde Betweenness
ceb<-cluster_edge_betweenness(friends)modularity(ceb)## [1] 0.5993285ceb$membership## [1] 1 1 2 2 1 1 1 1 2 2 1 1 1 1 2 1 2 2 1 1 1 3 1 2 4 1 2 4 4 2 2 3 4 2 4 1 1 4## [39] 2 2 2 3 3 3 3 3 3 3 3 3 3 3 3 5 3 5 5 5 5 5 5 5 5 5 5 5 5 5 5Comparing Clustering Algorithms
par(mfrow=c(2, 2), mar=c(0,0,1,0))plot(cfg, friends, layout=LO, main="Greedy")plot(wc, friends, layout=LO, main="Short Random Walks")plot(fg, friends, layout=LO,main= "Fast Greedy")plot(ceb, friends, layout=LO, main="Betweenness")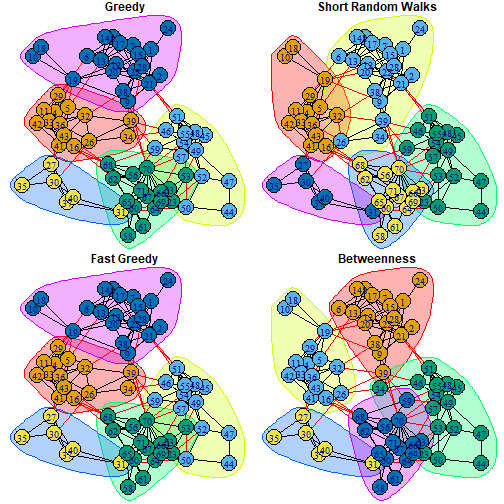
Your Turn
- Apply these algorithms to find communities in the email data (Assignment 1)
Challenge Yourself: Resume Data
As you were analyzing the email data, Tethys PD sent you the resume data that they were able to retrieve from GAStech servers. Scrape these resumes to identify each employee's education and previous employment. Use the officer packages to read each file and various stringr functions to extract the data you need. Then you can add the retrieved information to help identify additional connections among GAStech employees.
library(officer)myfiles<-list.files()doc <- officer::read_docx(path = myfiles[1])doc_text <- officer::docx_summary(doc)$textdoc_text <- paste(doc_text, collapse = " ")Helpful Functions from stringr
library(stringr)# Pattern to match everything after a specific word to a period.pattern <- paste0("Education","\\s(.*?)\\.")extracted_text <- str_extract(doc_text1,pattern)extracted_text# Pattern to match everything after a specific wordpattern <- paste0("Education", " .*")extracted_text <- str_extract(doc_text1, pattern)Write a Loop to Extract Name and Educ
Name<-NULLEduc<-NULLfor (i in 1:length(myfiles)){ doc <- officer::read_docx(path = myfiles[i]) doc_text <- officer::docx_summary(doc)$text Name[i]<-myfiles[i] |> str_remove("Bio")|> str_remove("Resume")|> str_remove(".docx") |> str_remove_all("[^A-Za-z0-9]") |> str_replace("(?<=\\p{Ll})(\\p{Lu})", " \\1") doc_text <- paste(doc_text, collapse = " ") if (str_detect(doc_text, "Education")==TRUE) { Educ[i]<-ifelse(str_detect(doc_text,"University of Tethys")==1,"University of Tethys", ifelse(str_detect(doc_text,"Tethys University")==1,"Tethys University", ifelse(str_detect(doc_text,"Abila Community College")==1,"Abila Community College", ifelse(str_detect(doc_text,"Barrington University")==1,"Barrington University", ifelse(str_detect(doc_text,"IT Tech University")==1,"IT Tech University",NA))))) } }mydata<-cbind.data.frame("Name"=Name, "Educ"=Educ)