Advanced Network Analysis
Random Graphs
Olga Chyzh [www.olgachyzh.com]
Why ERGMs?
The fundamental problem of network analysis:
Is the network we observe an Erdos-Renyi network?
- In an Erdos-Renyi network, the probability of each edge is independent of that of other edges.
If not, what are the endogeneous network features that define our network?
Example: Friendship Netwok
library(igraph)library(sna)data(coleman) #Use friendship datacoleman<-coleman[1,,]#convert to an -igraph- object, we'll treat it as a directed graph for now:coleman<-graph_from_adjacency_matrix(coleman, mode="directed", diag=FALSE) edge_density(coleman)## [1] 0.04623288reciprocity(coleman) #Note that -igraph- default is an undirected graph## [1] 0.5102881Friendship Data
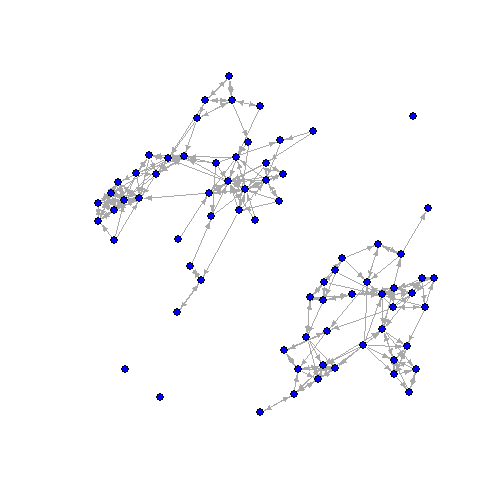
- What are some endogenous network features of this network?
Erdos-Renyi Networks
In order to answer whether an observed network is different from a random network, it would help to know what a random network would look like.
In a random network, all edges have the same probability of realization, p. Moreover, the probability of realization of edge i, pi, does not depend on pj.
Simulate A Random Graph to Compare to the Coleman Data
Need:
- number of nodes N in the friendship network
- the probability that each two nodes are connected p
vcount(coleman) #returns the number of vertices## [1] 73gsize(coleman) #returns the number of edges## [1] 243#orsummary(coleman)## IGRAPH fe3973e DN-- 73 243 -- ## + attr: name (v/c)Simulate A Random Graph to Compare to the Coleman Data
Let p denote the probability that any two vertices are connected by an edge. Then, we can calculate the value of p in the friendship network as the number of observated edges over the number of all possible edges.
With 73 vertices, we have 73∗72/2=2628 possible undirected edges or 73∗72=5256 directed edges, as each vertex can connect to each other vertex, but there are no self-loops. (Since the friendship network is directed, let's focus on simulating a directed network.)
Then p=243/5256=0.046. Does this value seem high or low? Note that p is the clustering coefficient of a random network with a given number of nodes and edges.
Simulate A Random Graph (Continued)
set.seed(45765) #since a simulation involves randomness, set the seed for #reproducibility.#Step i--start with a matrix of 73 unconnected nodes.N=73 #Set the number of nodes:rnet<-matrix(0, nrow=N,ncol=N) #Step ii:p<-243/5256for (i in 1:N) { for (j in 1:N){ if (i!=j) { rnet[i,j]=as.numeric(runif(1)<p) }}}Check Our Work
summary(g<-graph_from_adjacency_matrix(rnet, mode="directed", weighted=NULL))## IGRAPH fe59946 D--- 73 232 --edge_density(coleman)## [1] 0.04623288reciprocity(coleman) #Note that -igraph- default is an undirected graph## [1] 0.5102881edge_density(g)## [1] 0.04414003reciprocity(g) #Note that -igraph- default is an undirected graph## [1] 0.03448276Visualize
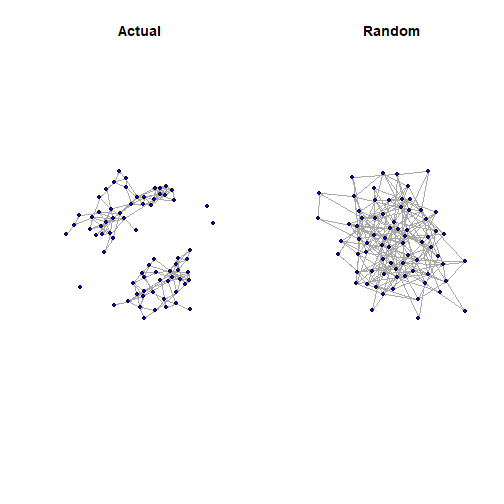
Summarize the Observed and Simulated Networks
library(statnet)data(coleman)coleman<- as.network.matrix(coleman[1,,], matrix.type='adjacency', directed=TRUE)rnet<- as.network.matrix(rnet, matrix.type='adjacency', directed=TRUE)summary(coleman~ edges+idegree(6)+ triangles+ mutual+ostar(2)+istar(2))## edges idegree6 triangle mutual ostar2 istar2 ## 243 5 460 62 383 542summary(rnet~edges+idegree(6)+ triangles+ mutual+ostar(2)+istar(2))## edges idegree6 triangle mutual ostar2 istar2 ## 232 5 37 4 343 345Your Turn
Simulate a random network that we could compare to the Sampson data.
Plot the two side-by-side
Use summary to further explore the differences between the two.
Based on this analysis, what model specification would you propose?