Advanced Network Analysis
Stochastic Actor-Oriented Models
Shahryar Minhas [s7minhas.com]
Why Use SOAM?
Until now, we modeled networks as a function of covariates (endogenous and exogenous).
Suppose we want to know whether network ties affect behavior? (e.g., friendships and bad habits)
Or whether ties in two different networks affect each other (e.g., work and romantic relationships)
SOAMs allow for modeling network(s) and nodal behavior as two mutually constitutive outcome variables.
Can you think of other examples of mutually-constitutive networks and behavior?
SAOM
Stochastic actor oriented model developed primarily by Snijders is implemented in the RSiena package on CRAN:
SAOM Assumptions
Actors control their outgoing ties and have full knowledge of the broader network
Changes in network ties and actor behavior happen in continuous time
Continuous time is modeled as a series of microsteps
During a microstep, a randomly chosen actor can change only one of its ties or the value of its behavior variable
Tie change only depends on the present network
SAOM Broadstrokes
The simulation starts out at the network observed at the first time point t0
- Examples: students in the same school
An actor is chosen randomly using a rate function
The identified actor gets the opportunity to set a micro step. The actor's choice is determined by their objective function
Model time is updated and simulation proceeds at step 2
The simulation terminates once modified network resembles network at t1
Rate Functions
Micro steps are opportunities for a randomly chosen actor to change one of his/her outgoing ties or behavior.
The frequency of these opportunities for change is modeled using rate functions, one for each type of change (network, behavior).
Waiting time between steps follows an exponential distribution with parameter λtN ( N refers to number of actors in the network)
Values of λt are estimated by calculating the number of edge differences between networks:
The higher λt is the greater the number of changes between observation moments
Probability that an actor i has the opportunity to make a change is equal to 1/N
Actor's Objective Function
Micro steps can be of two kinds: those that involve network changes or behavior changes
For network changes, the micro step consists of the change of one tie by a given actor.
Suppose x is the current network, and actor i has an opportunity to make a network change.
The next network x′ then must either equal to x or deviate from x by just one row element.
Hence, there are N possible outcomes, and i chooses the one that maximizes his/her utility function, called objective function, fneti(x,x′,z)+ϵneti(x,x′,z),
- where z is the current vector of behavior scores, fnet is a measure of actor's satisfaction with the result of the network decision, and ϵnet is the random component.
Multinomial Choice Model
- The choice probabilities can be expressed in multinomial logit shape:
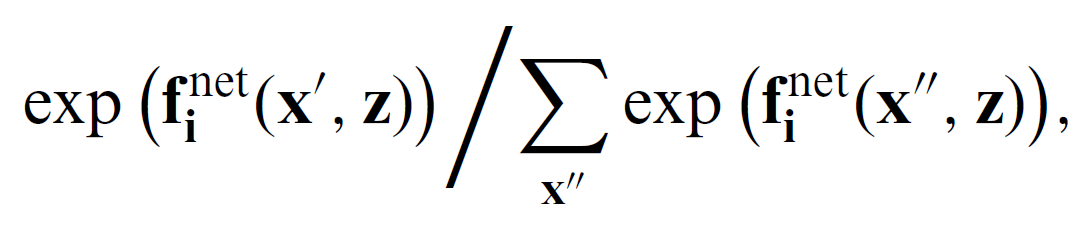
- where the sum in the denominator extends over all possible next network states x′′
Behavior Micro Steps
A given actor increments or decrements his score on the behavioral variable by one unit, provided that this change does not step outside the range of this variable. The score can also stay the same.
Let z denote the current vector of behavior scores for all actors, and z′ is the vector resulting from the actor's action in the micro step.
Actor i makes the choice by maximizing their behavior objective function, fbehi(x,z,z′)+ϵbehi(x,z,z′),
- where fbehi models the actor's satisfaction with the result of the behavior decision, and ϵbeh is the random error.
The behavior choice probability can be expressed as:
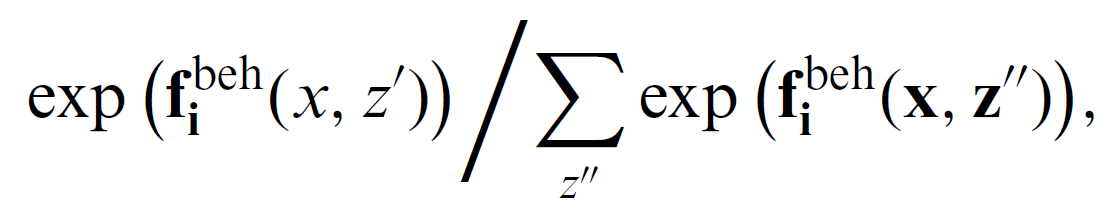
Parameterization
For network changes, the objective function has the general shape fneti(x,x′,z)=∑hβnethsneth(i,x,x′,z),
- where statistics sneth are the effects, weighted by parameters βneth.
For behavior changes:
fbehi(x,z,z′)=∑hβbehhsbehh(i,x,z,z′),
- Can specify exogenous and endogenous (network) effects.
Estimation
A stochastic simulation algorithm that generates network and behavioral data according to the postulated dynamic process.
Start with some network--behavior configuration, x(t), z(t).
Draw a waiting time and increment the parameter t by this waiting time.
Determine whether the next event is a network or a behavior change and select an actor who is making this change.
Determine the action of the selected actor.
Iterate until the end of the period is reached and evaluate the resulting simulated network--behavior configuration.
Example: Friendship Networks
library(RSiena) friend.data.w1 <- s501 friend.data.w2 <- s502 friend.data.w3 <- s503 drink <- s50a smoke <- s50sSpecify the Network DV:
friendship <- sienaDependent( array( c( friend.data.w1, friend.data.w2, friend.data.w3 ), dim = c( 50, 50, 3 ) ) )friendship## Type oneMode ## Observations 3 ## Nodeset Actors (50 elements)class(friendship)## [1] "sienaDependent"dim( friendship)## [1] 50 50 3attributes(friendship)## $dim## [1] 50 50 3## ## $class## [1] "sienaDependent"## ## $type## [1] "oneMode"## ## $sparse## [1] FALSE## ## $nodeSet## [1] "Actors"## ## $netdims## [1] 50 50 3## ## $allowOnly## [1] TRUESpecify the Behavior DV:
drinkingbeh <- sienaDependent( drink, type = "behavior" )drinkingbeh## Type behavior ## Observations 3 ## Nodeset Actors (50 elements)Specify IVs:
smoke1 <- coCovar( smoke[ , 1 ] )# Put the variables together in the data set for analysisNBdata <- sienaDataCreate( friendship, smoke1, drinkingbeh)NBdata## Dependent variables: friendship, drinkingbeh ## Number of observations: 3 ## ## Nodeset Actors ## Number of nodes 50 ## ## Dependent variable friendship ## Type oneMode ## Observations 3 ## Nodeset Actors ## Densities 0.046 0.047 0.05## ## Dependent variable drinkingbeh## Type behavior ## Observations 3 ## Nodeset Actors ## Range 1 - 5 ## ## Constant covariates: smoke1Possible Types of IVs
coCovar--constant node-level covariate (does not change between time periods)varCovar--time-variable node-level covariatecoDyadCovar--constant edge-level covariatevarDyadCovar--time-varying edge-level covariatesienaCompositionChange--over time changes in node set (e.g., some actors leave the network)
?coCovarSpecify Endogenous Effects
NBeff <- getEffects( NBdata )NBeff## name effectName include fix test ## 1 friendship constant friendship rate (period 1) TRUE FALSE FALSE## 2 friendship constant friendship rate (period 2) TRUE FALSE FALSE## 3 friendship outdegree (density) TRUE FALSE FALSE## 4 friendship reciprocity TRUE FALSE FALSE## 5 drinkingbeh rate drinkingbeh (period 1) TRUE FALSE FALSE## 6 drinkingbeh rate drinkingbeh (period 2) TRUE FALSE FALSE## 7 drinkingbeh drinkingbeh linear shape TRUE FALSE FALSE## 8 drinkingbeh drinkingbeh quadratic shape TRUE FALSE FALSE## initialValue parm## 1 4.69604 0 ## 2 4.32885 0 ## 3 -1.46770 0 ## 4 0.00000 0 ## 5 0.70571 0 ## 6 0.84939 0 ## 7 0.32237 0 ## 8 0.00000 0Effects Description
effectsDocumentation(NBeff)
Specify Effects
NBeff <- includeEffects( NBeff, transTrip, transRecTrip )## effectName include fix test initialValue parm## 1 transitive triplets TRUE FALSE FALSE 0 0 ## 2 transitive recipr. triplets TRUE FALSE FALSE 0 0NBeff <- includeEffects( NBeff, egoX, altX, altSqX, diffSqX, interaction1 = "smoke1" )## effectName include fix test initialValue parm## 1 smoke1 alter TRUE FALSE FALSE 0 0 ## 2 smoke1 squared alter TRUE FALSE FALSE 0 0 ## 3 smoke1 ego TRUE FALSE FALSE 0 0 ## 4 smoke1 diff. squared TRUE FALSE FALSE 0 0NBeff## name effectName include fix test ## 1 friendship constant friendship rate (period 1) TRUE FALSE FALSE## 2 friendship constant friendship rate (period 2) TRUE FALSE FALSE## 3 friendship outdegree (density) TRUE FALSE FALSE## 4 friendship reciprocity TRUE FALSE FALSE## 5 friendship transitive triplets TRUE FALSE FALSE## 6 friendship transitive recipr. triplets TRUE FALSE FALSE## 7 friendship smoke1 alter TRUE FALSE FALSE## 8 friendship smoke1 squared alter TRUE FALSE FALSE## 9 friendship smoke1 ego TRUE FALSE FALSE## 10 friendship smoke1 diff. squared TRUE FALSE FALSE## 11 drinkingbeh rate drinkingbeh (period 1) TRUE FALSE FALSE## 12 drinkingbeh rate drinkingbeh (period 2) TRUE FALSE FALSE## 13 drinkingbeh drinkingbeh linear shape TRUE FALSE FALSE## 14 drinkingbeh drinkingbeh quadratic shape TRUE FALSE FALSE## initialValue parm## 1 4.69604 0 ## 2 4.32885 0 ## 3 -1.46770 0 ## 4 0.00000 0 ## 5 0.00000 0 ## 6 0.00000 0 ## 7 0.00000 0 ## 8 0.00000 0 ## 9 0.00000 0 ## 10 0.00000 0 ## 11 0.70571 0 ## 12 0.84939 0 ## 13 0.32237 0 ## 14 0.00000 0Define the Model:
myalgorithm1 <- sienaAlgorithmCreate( projname = 's50_NB' )# Estimate using the second algorithm right from the start.NBans <- siena07(myalgorithm1, data = NBdata, effects = NBeff)NBans <- siena07(myalgorithm1, data = NBdata, effects = NBeff, batch=TRUE) #without Siena screenLook at results
NBans## Estimates, standard errors and convergence t-ratios## ## Estimate Standard Convergence ## Error t-ratio ## Network Dynamics ## 1. rate constant friendship rate (period 1) 6.2738 ( 1.2186 ) 0.0384 ## 2. rate constant friendship rate (period 2) 5.0845 ( 0.8572 ) -0.0337 ## 3. eval outdegree (density) -2.6246 ( 0.2222 ) 0.0538 ## 4. eval reciprocity 2.7737 ( 0.2664 ) 0.0710 ## 5. eval transitive triplets 0.8915 ( 0.1360 ) 0.1005 ## 6. eval transitive recipr. triplets -0.5130 ( 0.2160 ) 0.1082 ## 7. eval smoke1 alter 0.2548 ( 0.2914 ) -0.0221 ## 8. eval smoke1 squared alter -0.2197 ( 0.2489 ) 0.0047 ## 9. eval smoke1 ego 0.0873 ( 0.3034 ) 0.0015 ## 10. eval smoke1 squared ego 0.0104 ( 0.2501 ) 0.0322 ## 11. eval smoke1 diff. squared -0.0981 ( 0.0679 ) -0.0047 ## ## Behavior Dynamics## 12. rate rate drinkingbeh (period 1) 1.1712 ( 0.3042 ) -0.0364 ## 13. rate rate drinkingbeh (period 2) 1.6518 ( 0.4074 ) 0.0249 ## 14. eval drinkingbeh linear shape 0.3664 ( 0.1425 ) 0.0079 ## 15. eval drinkingbeh quadratic shape -0.2053 ( 0.0962 ) -0.0834 ## ## Overall maximum convergence ratio: 0.1885 ## ## ## Total of 3435 iteration steps.Interpretation of Parameters
The parameters on the rate functions are interpreted as related to the speed of the evolution process. These parameters reflect the frequencies of the opportunities for change.
The parameters on the objective functions may be interpreted in terms of their direction and statistical significance or by calculating predicted probabilities of ties in hypothetical scenarios (i.e., use the reverse logistic transformation).
The linear and the quadratic shape parameters of the behavior objective function model the shape of the long-term distribution of the behavior variable.
Can also use the reverse logit transformation to interpret the parameters on the behavior effects.
Example
Suppose we estimated the following model:
fneti=−2∑jx′ij+2.5∑jx′ijxji+1∑jx′ijsimij
and fbehi(x,z,z′)=−1(z′i−z)−.5(z′i−z)2+2.5(∑jxijsim′ij)/(∑jxij)
The primes indicate the elements in the formulae that are under control of actor i and may be changed in a micro step.
- Note that the network objective function contains outdegree, reciprocity, and the similarity effect, and the behavior function includes the linear and quadratic shape of the distribution of the behavior variable and an effect of average similarity to neighbors.
Interpretation of Parameters: Network Function
The parameter estimate of -2 on the outdegree tells us that, if we ignore all other variables, the baseline probability of a tie in this network is exp(−2)/(1+exp(−2))=0.12.
The parameter estimate of 2.5 on the reciprocity tells us that the probability of a reciprocated tie is exp(−2+2.5)/(1+exp(−2+2.5))=0.62.
The positive similarity effect indicates that actors tend to form ties with similar others rather than to dissimilar ones. The probability of a tie to a maximally similar actor is exp(−2+1)/(1+exp(−2+1))=0.26 vs that of the baseline probability of 0.12.
Interpretation of Parameters: Behavior Function
Suppose our behavior variable ranges from 1 to 5 and has an average of 3. When mean centered, this variable then ranges from -2 to 2.
The first two parameters define a parabolic shape. We can calculate the inflection point of this parabola by solving −1−0.5∗2x=0 for x. The result is x=-1, which corresponds to the value of 2 on the original scale of this variable. This means that in the long run, the distribution of the behavior variable is unimodal with a maximum at score value 2.
Can also calculate the probability of moving from one score to the other. For example, the probability of moving from 3 to 2 is exp(−1(2−3)−0.5(2−3)2)/(1+exp(−1(2−3)−0.5(2−3)2))=0.62 The probability of staying at 3 rather than moving to 2 is 1-0.62=0.38.
Interpretation of Parameters: Behavior Function Cont'd
The third parameter of 2.5 on the average similarity indicates that actors tend to act in the same manner as their friends.
Consider an actor with a 3 on their behavior and assume he has five friends, four of whom score 3 and 1 scoring 2 or lower.
If this actor moves to 2, his average similarity to four of his friends will decrease by -1*4 and his similarity to his fifth friend will increase by 1.
The average similarity will decrease from 1/5 to 4/5 for a change of -0.6. Then, accounting for the similarity effect, the probability of moving from 3 to 2 for this actor is exp(−1(2−3)−0.5(2−3)2+2.5(−0.6))/(1+exp(−1(2−3)−0.5(2−3)2+2.5(−0.6))=0.26
Let's Specify a Model That...
- accounts for homophily effect for smoking
NBeff <- getEffects( NBdata )NBeff <- includeEffects(NBeff, simX, interaction1 = "smoke1" )## effectName include fix test initialValue parm## 1 smoke1 similarity TRUE FALSE FALSE 0 0Let's Specify a Model That...
- Parses out whether there is a selection or influence (or both) effect for drinking behavior--- include sender, receiver and homophily effects of drinking for friendship formation, and vice versa.
NBeff <- includeEffects(NBeff, egoX, altX, simX, interaction1 = "drinkingbeh" )## effectName include fix test initialValue parm## 1 drinkingbeh alter TRUE FALSE FALSE 0 0 ## 2 drinkingbeh ego TRUE FALSE FALSE 0 0 ## 3 drinkingbeh similarity TRUE FALSE FALSE 0 0NBeff <- includeEffects(NBeff, name = "drinkingbeh", avAlt,indeg,outdeg, interaction1 = "friendship" )## effectName include fix test initialValue parm## 1 drinkingbeh indegree TRUE FALSE FALSE 0 0 ## 2 drinkingbeh outdegree TRUE FALSE FALSE 0 0 ## 3 drinkingbeh average alter TRUE FALSE FALSE 0 0Results
## Estimates, standard errors and convergence t-ratios## ## Estimate Standard Convergence ## Error t-ratio ## Network Dynamics ## 1. rate constant friendship rate (period 1) 5.9052 ( 0.9341 ) -0.0012 ## 2. rate constant friendship rate (period 2) 4.5123 ( 0.7046 ) -0.0224 ## 3. eval outdegree (density) -2.4804 ( 0.1415 ) 0.0008 ## 4. eval reciprocity 2.7742 ( 0.2659 ) 0.0052 ## 5. eval smoke1 similarity 0.2967 ( 0.2100 ) 0.0154 ## 6. eval drinkingbeh alter -0.0144 ( 0.1147 ) -0.0027 ## 7. eval drinkingbeh ego 0.1293 ( 0.1177 ) 0.0406 ## 8. eval drinkingbeh similarity 1.3202 ( 0.6778 ) -0.0124 ## ## Behavior Dynamics## 9. rate rate drinkingbeh (period 1) 1.2619 ( 0.3548 ) 0.0043 ## 10. rate rate drinkingbeh (period 2) 1.7245 ( 0.4400 ) 0.0430 ## 11. eval drinkingbeh linear shape -1.2946 ( 2.8020 ) 0.0164 ## 12. eval drinkingbeh quadratic shape -1.5840 ( 2.6522 ) -0.0539 ## 13. eval drinkingbeh indegree -0.8568 ( 2.1755 ) 0.0035 ## 14. eval drinkingbeh outdegree 1.7257 ( 3.6961 ) 0.0038 ## 15. eval drinkingbeh average alter 3.3212 ( 5.7404 ) -0.0847 ## ## Overall maximum convergence ratio: 0.1387 ## ## ## Total of 3266 iteration steps.## ## Covariance matrix of estimates (correlations below diagonal)## ## 0.872 0.052 -0.007 0.050 0.013 0.005 0.005 -0.208 -0.016 -0.023 -0.728 -0.819 -0.588 1.066 1.787## 0.079 0.497 0.000 0.014 0.007 0.006 -0.003 -0.096 -0.020 -0.010 -0.475 -0.426 -0.297 0.572 0.902## -0.052 0.001 0.020 -0.028 -0.006 0.001 -0.004 -0.009 0.004 -0.008 0.140 0.172 0.147 -0.243 -0.370## 0.201 0.077 -0.734 0.071 0.007 0.000 0.008 -0.070 -0.016 0.007 -0.371 -0.447 -0.395 0.649 0.960## 0.067 0.048 -0.195 0.121 0.044 0.004 0.006 -0.023 -0.003 0.001 -0.044 -0.061 -0.064 0.095 0.129## 0.045 0.074 0.043 0.000 0.181 0.013 -0.008 -0.015 -0.004 0.002 -0.087 -0.064 -0.060 0.105 0.135## 0.047 -0.041 -0.264 0.255 0.243 -0.559 0.014 -0.005 -0.002 0.002 -0.007 -0.025 -0.024 0.035 0.050## -0.329 -0.200 -0.099 -0.386 -0.161 -0.191 -0.058 0.459 0.058 0.035 0.920 0.967 0.776 -1.365 -2.086## -0.049 -0.080 0.070 -0.169 -0.041 -0.087 -0.052 0.239 0.126 0.000 0.219 0.225 0.231 -0.383 -0.473## -0.056 -0.032 -0.126 0.059 0.012 0.044 0.042 0.117 -0.003 0.194 0.032 -0.022 -0.071 0.051 0.045## -0.278 -0.240 0.353 -0.498 -0.075 -0.270 -0.021 0.484 0.221 0.026 7.851 6.551 4.669 -9.236 -13.952## -0.330 -0.228 0.458 -0.634 -0.109 -0.209 -0.081 0.538 0.239 -0.019 0.882 7.034 5.399 -9.578 -15.162## -0.289 -0.194 0.477 -0.684 -0.140 -0.240 -0.092 0.526 0.299 -0.074 0.766 0.936 4.733 -7.813 -11.565## 0.309 0.220 -0.465 0.660 0.123 0.247 0.081 -0.545 -0.292 0.031 -0.892 -0.977 -0.972 13.661 20.501## 0.333 0.223 -0.456 0.629 0.107 0.205 0.073 -0.536 -0.232 0.018 -0.867 -0.996 -0.926 0.966 32.953## ## Derivative matrix of expected statistics X by parameters:## ## 10.492 0.000 1.260 1.621 0.022 -0.286 -0.036 0.360 -0.014 0.000 -0.101 0.435 -0.787 -0.893 -1.038## 0.000 12.230 0.494 1.004 -0.260 0.793 -0.108 -0.167 0.000 0.296 0.661 1.854 1.714 0.559 0.506## 20.513 22.083 209.478 159.357 20.998 -13.092 -13.352 16.852 6.532 5.113 30.650 10.932 70.858 77.206 6.519## 1.156 3.194 90.373 107.430 8.589 -3.697 -8.335 8.454 1.149 1.122 11.879 1.973 31.667 29.911 4.338## 1.803 1.867 21.328 15.793 32.161 -34.775 -29.411 3.529 -0.502 0.244 0.894 0.310 4.853 7.776 -1.215## 0.867 1.407 10.176 20.746 -26.217 199.379 124.634 1.248 3.125 0.522 32.973 15.985 66.945 58.831 15.883## 4.936 8.254 14.044 2.225 -29.707 130.336 182.602 -0.179 0.043 -5.478 2.915 25.764 5.475 3.668 19.069## 4.979 4.274 24.592 20.679 3.378 1.331 0.366 8.337 0.104 -0.509 4.282 5.433 9.490 10.157 2.655## -1.539 0.000 -1.036 -1.106 -0.206 -0.480 -0.401 -1.965 14.198 0.000 8.809 5.557 18.751 21.167 4.112## 0.000 0.556 1.226 0.588 0.252 -2.818 -3.108 -1.013 0.000 11.461 6.660 0.697 18.187 16.110 5.948## -0.652 2.394 3.863 2.155 0.256 3.593 3.278 0.103 2.622 3.112 26.295 9.288 59.100 59.793 8.256## -2.814 -1.390 -10.149 -4.709 -2.422 6.046 7.378 0.254 0.971 0.855 2.985 79.375 18.974 18.009 46.333## -1.544 4.252 10.994 9.329 1.662 6.530 7.034 0.904 5.770 9.381 66.415 29.782 173.983 164.298 23.472## -1.896 4.502 6.699 3.974 0.858 3.492 5.362 0.903 7.079 9.315 60.959 37.344 153.726 154.403 19.158## -1.008 -0.778 -2.283 -0.737 -0.742 3.695 3.249 0.293 -0.551 -0.888 -1.759 27.900 -0.200 -2.861 21.845## ## Covariance matrix of X (correlations below diagonal):## ## 88.047 4.936 34.813 19.504 3.615 -5.717 0.060 3.486 0.474 -1.546 3.934 -2.844 8.710 3.289 -6.759## 0.061 74.490 26.993 12.060 3.014 -3.639 -5.094 1.320 -0.538 2.203 5.610 -1.956 11.041 10.738 -4.924## 0.258 0.217 207.589 162.636 21.136 -15.785 -13.095 16.074 6.885 4.360 37.538 -11.499 81.884 86.531 -3.814## 0.155 0.104 0.841 180.171 15.389 -1.646 -8.420 15.901 5.562 3.104 31.470 3.190 71.355 74.122 5.015## 0.067 0.060 0.254 0.198 33.460 -37.731 -35.510 3.950 0.340 -0.324 3.211 -3.343 10.301 10.870 -1.991## -0.038 -0.027 -0.069 -0.008 -0.411 252.455 165.576 -0.148 -1.181 -1.902 13.812 31.600 16.507 15.351 24.739## 0.000 -0.038 -0.058 -0.040 -0.394 0.670 242.220 -1.038 -4.925 -7.963 4.775 38.527 5.080 2.621 25.651## 0.101 0.042 0.303 0.321 0.185 -0.003 -0.018 13.583 -1.804 -1.266 2.856 7.705 8.218 6.946 2.872## 0.011 -0.014 0.104 0.090 0.013 -0.016 -0.069 -0.106 21.253 -0.565 13.496 1.530 26.099 31.801 2.199## -0.033 0.051 0.061 0.046 -0.011 -0.024 -0.103 -0.069 -0.025 24.815 14.865 -0.121 39.065 38.423 0.947## 0.044 0.068 0.271 0.244 0.058 0.090 0.032 0.081 0.304 0.310 92.532 21.368 221.627 216.475 32.788## -0.021 -0.016 -0.056 0.017 -0.040 0.139 0.173 0.146 0.023 -0.002 0.156 204.031 85.787 67.568 131.808## 0.036 0.049 0.220 0.205 0.069 0.040 0.013 0.086 0.219 0.303 0.890 0.232 669.684 582.166 100.183## 0.014 0.051 0.245 0.225 0.077 0.039 0.007 0.077 0.282 0.315 0.919 0.193 0.919 599.874 68.499## -0.056 -0.044 -0.021 0.029 -0.027 0.121 0.128 0.061 0.037 0.015 0.265 0.717 0.301 0.217 165.817Convergence Ratios
Note that the "convergence t-ratio" is the t-ratio for convergence checking, not the t statistic for testing the significance of this effect (See Section 6.3 of the RSiena manual).
For good convergence, the t-ratios for convergence all should be less than .1 in absolute value, and the overall maximum convergence ratio should be less than 0.25.
If this is not yet the case, you should try again, starting from the last estimate as the previous answer (
prevAnsargument), e.g.:
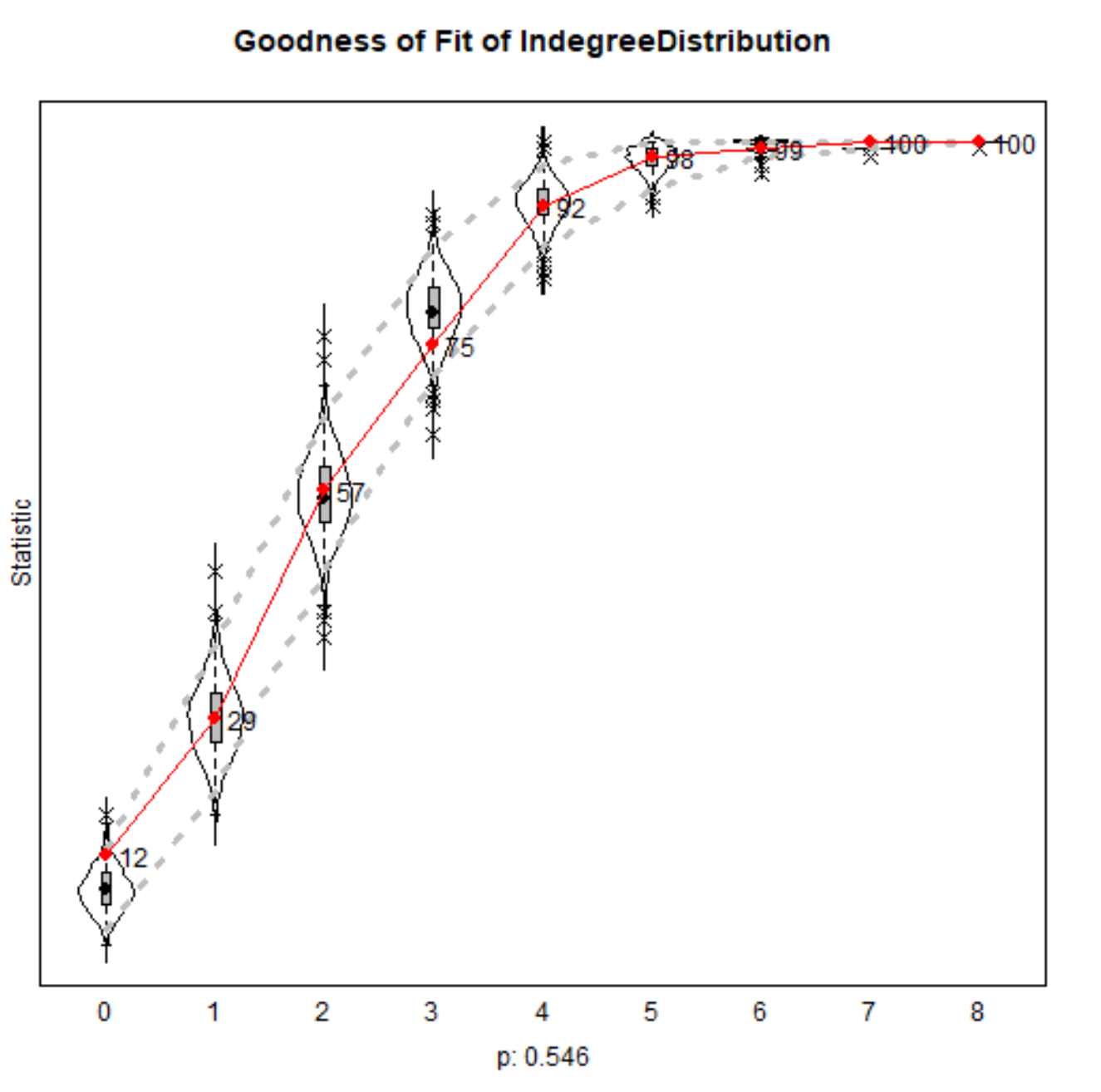
m3 <- siena07(myalgorithm2, data = NBdata, effects = NBeff, prevAns = m2 )Goodness of Fit: Indegree
Can plot distributions of network statistics from simulated networks
Note: must specify
returnDeps=TRUEin thesiena07function to save these networks.
myalgorithm2 <- sienaAlgorithmCreate(projname = 's50CoEv_2' )m2 <- siena07(myalgorithm2, data = NBdata, effects = NBeff, batch=TRUE, returnDeps=TRUE) gofI <- sienaGOF(m2, IndegreeDistribution, verbose=TRUE, join=TRUE, varName="friendship")plot(gofI)saveRDS(gofI,"./data/gofI")Goodness of Fit: Indegree
Goodness of Fit: Outdegree
gofO <- sienaGOF(m2, OutdegreeDistribution, verbose=TRUE, join=TRUE, varName="friendship")plot(gofO)saveRDS(gofO,"data/gofO")
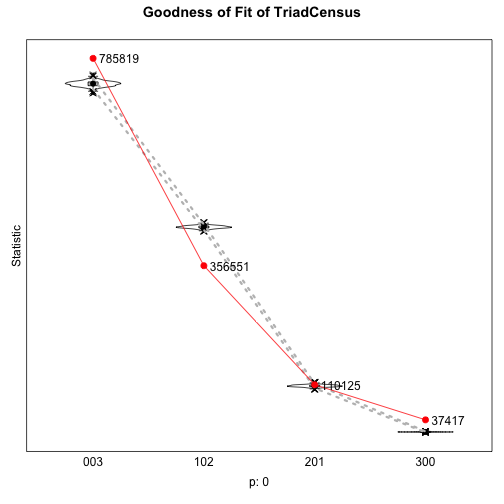
Goodness of Fit: Triad Census
gofTC <- sienaGOF(m2, TriadCensus, verbose=TRUE, join=TRUE, varName="friendship")plot(gofTC)saveRDS(gofTC,"data/gofTC")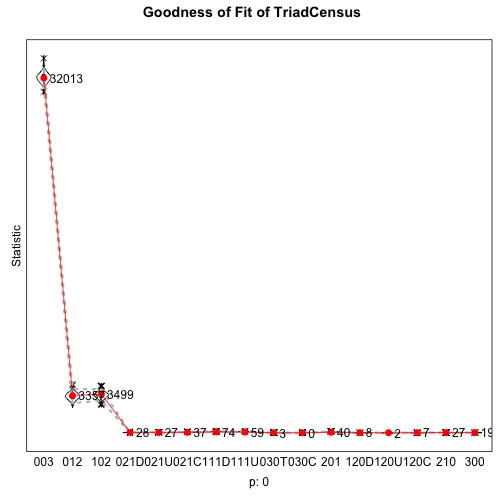
Example: Knecht Data
library(btergm)data(knecht)Longitudinal classroom friendship network and behavior (Andrea Knecht)
friendship--a list of adjacency matrices at four time periodsdemographics--node-level covariates: sex, age, ethnicity, religiondelinquency--number of delinquencies
Actors Entering and Exiting the Network
Treatment by structural zero coding
Treatment by composition change directives
What to use?
Loosely related to structural zeros: structural ones
Treatment by structural zero coding
When actors are not part of the group at a given measurement point, code their outgoing and incoming ties as “10”, meaning “absent, and could not possibly have been present”.
When running simulations, this is handled as follows:
- A tie value “10” at the beginning of a period implies that the tie will remain structurally absent throughout the period, no matter what the tie’s value at the end of the period is.
- A tie value “10” at the end of a period implies that no matter what the tie’s simulated value at the end of the period is, it is overwritten by “10” before any statistics are evaluated.
See RSiena manual section 4.1.2.
Treatment by structural zero coding
When information is known about the exact time when actors left or entered the group in continuous time between observation moments, this information can be made use of.
In simulations, joiners enter at the indicated time point and then are simultaneously connected to the rest of the actors according to the data provided for the period begin (so, they do not necessarily have to ‘start from scratch’ but can inherit ties!)
Leavers just exit and cannot change their ties any more from this time point on; their last connection data can be provided for the period end.
Joiner and leaver data need to be provided in an additional file; see RSiena manual sections 2.1.2 and 4.7.
What to Use
Composition change directives allow to make use of more information. If information is scarce, this may be the better option.
Structural zero treatment is quite crude, if results can be obtained this way, they will likely be robust. But under scarce information conditions, it can happen that no results can be obtained.
Structural Ones
Sometimes, ties can be “present, and could not possibly have been absent”.
Studying a communication network among employees, where Loosely related to structural zeros:
Studying a communication network among employees, where some people are forced to communicate anyway (by their job contract).
Studying a growing network where ties once formed cannot be dissolved again.
In such situations, tie variables can be coded as “11”.
See RSiena manual section 4.1.2.
Example: Duque Data
library(devtools)#install_github("ochyzh/networkdata")data("duqueData")dim(dipl_ties[[1]])## [1] 134 134dim(dipl_ties[[2]])## [1] 148 148Remember that in these data, time periods have varying numbers of observations, as states enter and leave the system.
In order to use RSiena, we must have the same number of actors in each time period. If an actor is missing, their tie values are coded using the structural zero code
10. Alternatively, you can provide an additional file that details when actors enter and leave the data.
Example: Duque Data
library(tidyverse)#get the full list of actors:myactors<-sort(as.numeric(unique(do.call("c",lapply(dipl_ties[1:3],names)))))n<-length(myactors)dipl<-array(10, dim = c(n,n, 3 ), dimnames=list(myactors,myactors,seq(from=1970,to=1980,by=5)))dipl[1:10,1:10,1]for(t in 1:3){ d<-dipl_ties[[t]] for(i in 1:nrow(d)){ for (j in 1:ncol(d)){ a1 = names(d)[i] a2 = colnames(d)[j] val = as.numeric(as.character(d[i,j])) dipl[i,j,t] <- val dipl[j,i,t] <- val }} }dipl <- sienaDependent(dipl)Results
## Estimates, standard errors and convergence t-ratios## ## Estimate Standard Convergence ## Error t-ratio ## ## Rate parameters: ## 0.1 Rate parameter period 1 59.0072 ( 3.7459 ) ## 0.2 Rate parameter period 2 55.7625 ( 2.8318 ) ## ## Other parameters: ## 1. eval degree (density) -0.3857 ( 0.3191 ) -0.0117 ## 2. eval transitive ties -0.2815 ( 0.3132 ) -0.0083 ## 3. eval cont 0.6214 ( 0.0600 ) -0.0381 ## 4. eval ally 0.9287 ( 0.0347 ) 0.0433 ## 5. eval dem similarity -0.3061 ( 0.0221 ) 0.0352 ## ## Overall maximum convergence ratio: 0.1007 ## ## ## Total of 2267 iteration steps.## ## Covariance matrix of estimates (correlations below diagonal)## ## 0.102 -0.100 -0.001 0.000 -0.002## -1.000 0.098 0.001 0.000 0.002## -0.053 0.048 0.004 0.000 0.000## 0.019 -0.024 -0.111 0.001 0.000## -0.257 0.260 -0.075 -0.074 0.000## ## Derivative matrix of expected statistics X by parameters:## ## 5902.370 12067.765 89.267 313.469 -512.411## 6025.170 12339.766 87.772 320.229 -569.246## 127.614 263.295 329.969 80.854 75.560## 253.418 504.793 91.289 1030.137 12.066## -285.863 -596.013 73.841 98.099 2442.961## ## Covariance matrix of X (correlations below diagonal):## ## 3283.527 6735.999 5.567 150.540 -365.027## 0.998 13861.069 15.914 293.853 -761.672## 0.005 0.007 377.997 110.570 82.376## 0.075 0.071 0.162 1225.602 1.596## -0.123 -0.125 0.082 0.001 2678.907Goodness of Fit: Outdegree
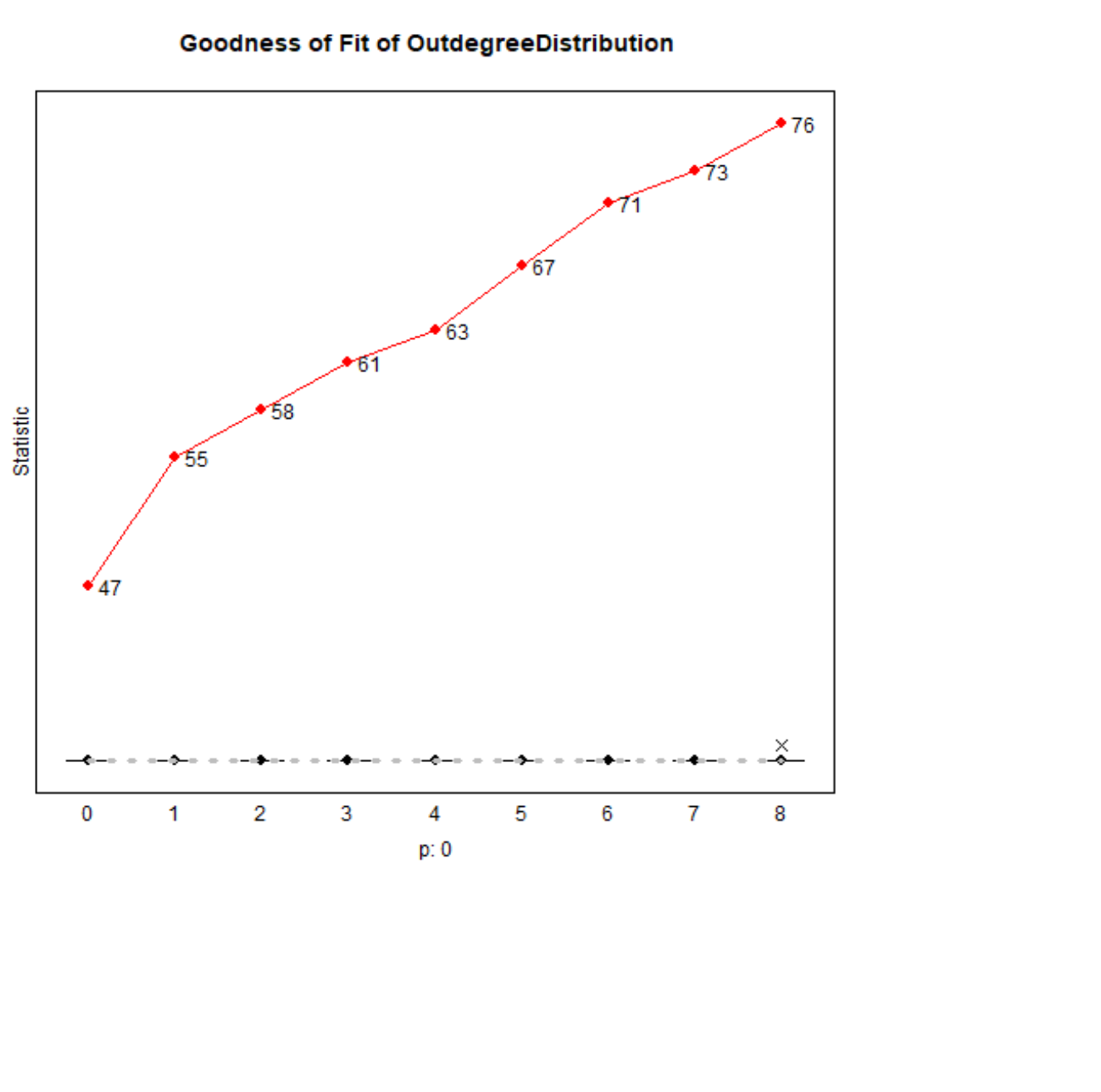
Goodness of Fit: Triad Census
gofTC_dipl <- sienaGOF(ans, TriadCensus, verbose=TRUE, join=TRUE, varName="dipl")plot(gofTC_dipl)## Note: some statistics are not plotted because their variance is 0.## This holds for the statistics: 012 021D 021U 021C 111D 111U 030T 030C 120D 120U 120C 210.